-
SPSS 스타일로 데이터 분석하기R 2018. 12. 27. 19:00
유용한 패키지를 발견했다.
마케팅 분야나 사회조사에서 빈도표와 교차표(분할표)를 아주 많이 사용하고 있다. 얼마전에 강의했던 곳에서도 SPSS를 대체해서 R로 데이터분석을 하고 싶은데, 가장 많은 질문은 SPSS로 작성하는 빈도표나 교차표를 어떻게 R에서 작성하는 지를 많이 질문했다. 또한 변수가 가지는 값에 대한 설명의 설정도 질문했다.
이 질문에 대답이 될 수 있는 하나의 패키지이다.
바로 expss 이다.아래의 사이트를 참고하면 많은 도움이 될 것 같다.
https://gdemin.github.io/expss/앞으로 이 부분을 잘 학습해서,
마케팅 관련된 분야에서 근무하시는 사람들을 위한 책이나 강의를 해야겠다.expss: Tables with Labels in R
2018-11-12
Introduction
expsspackage provides tabulation functions with support for ‘SPSS’-style labels, multiple / nested banners, weights, multiple-response variables and significance testing. There are facilities for nice output of tables in ‘knitr’, R notebooks, ‘Shiny’ and ‘Jupyter’ notebooks. Proper methods for labelled variables add value labels support to base R functions and to some functions from other packages. Additionally, the package offers useful functions for data processing in marketing research / social surveys - popular data transformation functions from ‘SPSS’ Statistics (‘RECODE’, ‘COUNT’, ‘COMPUTE’, ‘DO IF’, etc.) and ‘Excel’ (‘COUNTIF’, ‘VLOOKUP’, etc.). Package is intended to help people to move data processing from ‘Excel’/‘SPSS’ to R. See examples below. You can get help about any function by typing?function_namein the R console.Installation
expssis on CRAN, so for installation you can print in the consoleinstall.packages("expss").Cross-tablulation examples
We will use for demonstartion well-known
mtcarsdataset. Let’s start with adding labels to the dataset. Then we can continue with tables creation.library(expss) data(mtcars) mtcars = apply_labels(mtcars, mpg = "Miles/(US) gallon", cyl = "Number of cylinders", disp = "Displacement (cu.in.)", hp = "Gross horsepower", drat = "Rear axle ratio", wt = "Weight (1000 lbs)", qsec = "1/4 mile time", vs = "Engine", vs = c("V-engine" = 0, "Straight engine" = 1), am = "Transmission", am = c("Automatic" = 0, "Manual"=1), gear = "Number of forward gears", carb = "Number of carburetors" )For quick cross-tabulation there are
freandcrofamily of function. For simplicity we demonstrate here onlycro_cpctwhich caluclates column percent. Documentation for other functions, such ascro_casesfor counts,cro_rpctfor row percent,cro_tpctfor table percent andcro_funfor custom summary functions can be seen by typing?croand?cro_funin the console.# 'cro' examples # just simple crosstabulation, similar to base R 'table' function cro(mtcars$am, mtcars$vs)Engine V-engine Straight engine Transmission Automatic 12 7 Manual 6 7 #Total cases 18 14 # Table column % with multiple banners cro_cpct(mtcars$cyl, list(total(), mtcars$am, mtcars$vs))#Total Transmission Engine Automatic Manual V-engine Straight engine Number of cylinders 4 34.4 15.8 61.5 5.6 71.4 6 21.9 21.1 23.1 16.7 28.6 8 43.8 63.2 15.4 77.8 #Total cases 32 19 13 18 14 # or, the same result with another notation mtcars %>% calc_cro_cpct(cyl, list(total(), am, vs))#Total Transmission Engine Automatic Manual V-engine Straight engine Number of cylinders 4 34.4 15.8 61.5 5.6 71.4 6 21.9 21.1 23.1 16.7 28.6 8 43.8 63.2 15.4 77.8 #Total cases 32 19 13 18 14 # Table with nested banners (column %). mtcars %>% calc_cro_cpct(cyl, list(total(), am %nest% vs))#Total Transmission Automatic Manual Engine Engine V-engine Straight engine V-engine Straight engine Number of cylinders 4 34.4 42.9 16.7 100 6 21.9 57.1 50.0 8 43.8 100 33.3 #Total cases 32 12 7 6 7 We have more sophisticated interface for table construction with
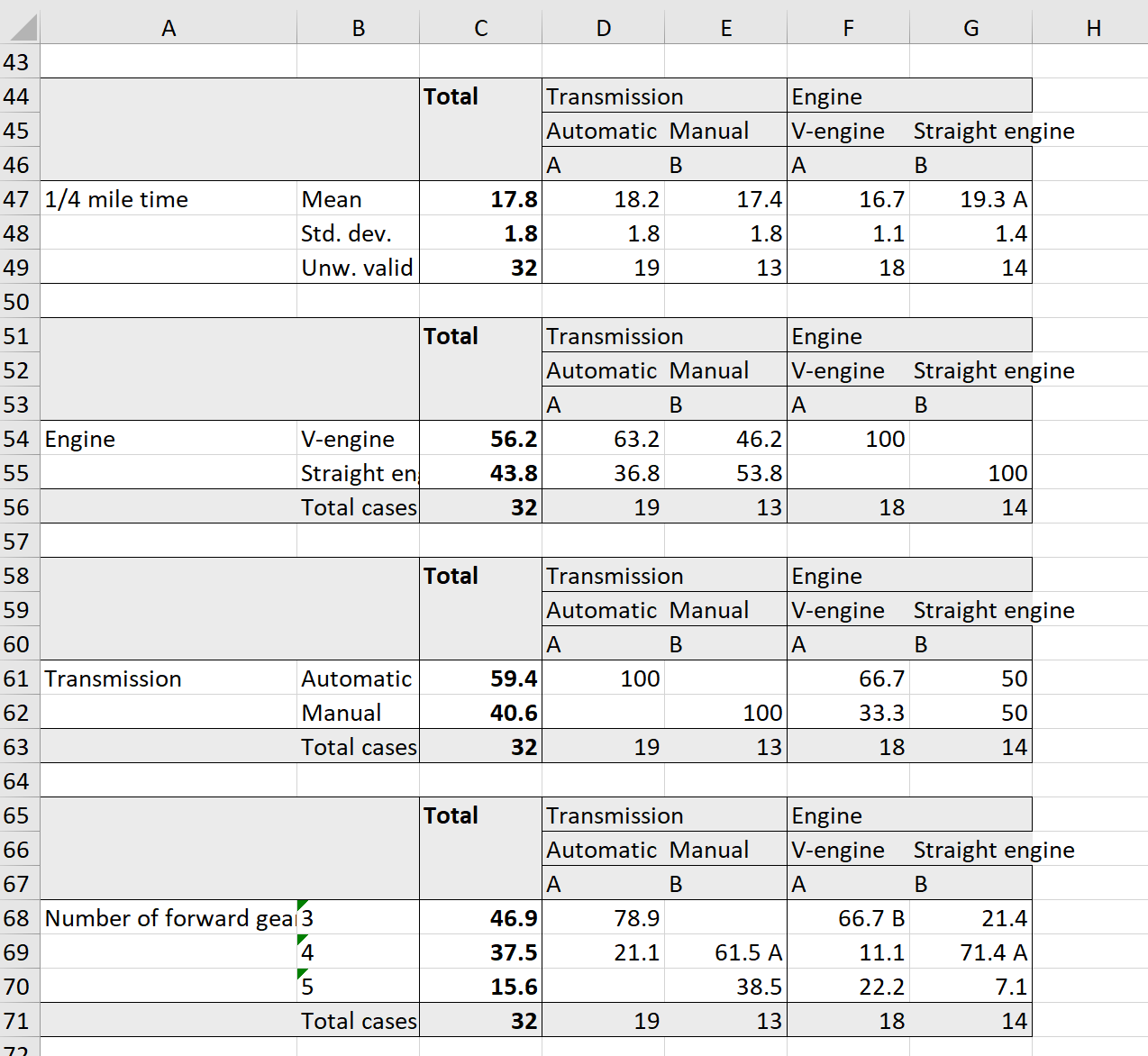
magrittrpiping. Table construction consists of at least of three functions chained with pipe operator:%>%. At first we need to specify variables for which statistics will be computed withtab_cells. Secondary, we calculate statistics with one of thetab_stat_*functions. And last, we finalize table creation withtab_pivot, e. g.:dataset %>% tab_cells(variable) %>% tab_stat_cases() %>% tab_pivot(). After that we can optionally sort table withtab_sort_asc, drop empty rows/columns withdrop_rcand transpose withtab_transpose. Resulting table is just adata.frameso we can use usual R operations on it. Detailed documentation for table creation can be seen via?tables. For significance testing see?significance. Generally, tables automatically translated to HTML for output in knitr or Jupyter notebooks. However, if we want HTML output in the R notebooks or in the RStudio viewer we need to set options for that:expss_output_rnotebook()orexpss_output_viewer().# simple example mtcars %>% tab_cells(cyl) %>% tab_cols(total(), am) %>% tab_stat_cpct() %>% tab_pivot()#Total Transmission Automatic Manual Number of cylinders 4 34.4 15.8 61.5 6 21.9 21.1 23.1 8 43.8 63.2 15.4 #Total cases 32 19 13 # table with caption mtcars %>% tab_cells(mpg, disp, hp, wt, qsec) %>% tab_cols(total(), am) %>% tab_stat_mean_sd_n() %>% tab_last_sig_means(subtable_marks = "both") %>% tab_pivot() %>% set_caption("Table with summary statistics and significance marks.")Table with summary statistics and significance marks. #Total Transmission Automatic Manual A B Miles/(US) gallon Mean 20.1 17.1 < B 24.4 > A Std. dev. 6.0 3.8 6.2 Unw. valid N 32.0 19.0 13.0 Displacement (cu.in.) Mean 230.7 290.4 > B 143.5 < A Std. dev. 123.9 110.2 87.2 Unw. valid N 32.0 19.0 13.0 Gross horsepower Mean 146.7 160.3 126.8 Std. dev. 68.6 53.9 84.1 Unw. valid N 32.0 19.0 13.0 Weight (1000 lbs) Mean 3.2 3.8 > B 2.4 < A Std. dev. 1.0 0.8 0.6 Unw. valid N 32.0 19.0 13.0 1/4 mile time Mean 17.8 18.2 17.4 Std. dev. 1.8 1.8 1.8 Unw. valid N 32.0 19.0 13.0 # Table with the same summary statistics. Statistics labels in columns. mtcars %>% tab_cells(mpg, disp, hp, wt, qsec) %>% tab_cols(total(label = "#Total| |"), am) %>% tab_stat_fun(Mean = w_mean, "Std. dev." = w_sd, "Valid N" = w_n, method = list) %>% tab_pivot()#Total Transmission Automatic Manual Mean Std. dev. Valid N Mean Std. dev. Valid N Mean Std. dev. Valid N Miles/(US) gallon 20.1 6.0 32 17.1 3.8 19 24.4 6.2 13 Displacement (cu.in.) 230.7 123.9 32 290.4 110.2 19 143.5 87.2 13 Gross horsepower 146.7 68.6 32 160.3 53.9 19 126.8 84.1 13 Weight (1000 lbs) 3.2 1.0 32 3.8 0.8 19 2.4 0.6 13 1/4 mile time 17.8 1.8 32 18.2 1.8 19 17.4 1.8 13 # Different statistics for different variables. mtcars %>% tab_cols(total(), vs) %>% tab_cells(mpg) %>% tab_stat_mean() %>% tab_stat_valid_n() %>% tab_cells(am) %>% tab_stat_cpct(total_row_position = "none", label = "col %") %>% tab_stat_rpct(total_row_position = "none", label = "row %") %>% tab_stat_tpct(total_row_position = "none", label = "table %") %>% tab_pivot(stat_position = "inside_rows")#Total Engine V-engine Straight engine Miles/(US) gallon Mean 20.1 16.6 24.6 Valid N 32.0 18.0 14.0 Transmission Automatic col % 59.4 66.7 50.0 row % 100.0 63.2 36.8 table % 59.4 37.5 21.9 Manual col % 40.6 33.3 50.0 row % 100.0 46.2 53.8 table % 40.6 18.8 21.9 # Table with split by rows and with custom totals. mtcars %>% tab_cells(cyl) %>% tab_cols(total(), vs) %>% tab_rows(am) %>% tab_stat_cpct(total_row_position = "above", total_label = c("number of cases", "row %"), total_statistic = c("u_cases", "u_rpct")) %>% tab_pivot()#Total Engine V-engine Straight engine Transmission Automatic Number of cylinders #number of cases 19 12 7 #row % 100 63.2 36.8 4 15.8 42.9 6 21.1 57.1 8 63.2 100.0 Manual Number of cylinders #number of cases 13 6 7 #row % 100 46.2 53.8 4 61.5 16.7 100.0 6 23.1 50.0 8 15.4 33.3 # Linear regression by groups. mtcars %>% tab_cells(sheet(mpg, disp, hp, wt, qsec)) %>% tab_cols(total(label = "#Total| |"), am) %>% tab_stat_fun_df( function(x){ frm = reformulate(".", response = names(x)[1]) model = lm(frm, data = x) sheet('Coef.' = coef(model), confint(model) ) } ) %>% tab_pivot()#Total Transmission Automatic Manual Coef. 2.5 % 97.5 % Coef. 2.5 % 97.5 % Coef. 2.5 % 97.5 % (Intercept) 27.3 9.6 45.1 21.8 -1.9 45.5 13.3 -21.9 48.4 Displacement (cu.in.)0.0 0.0 0.0 0.0 0.0 0.0 0.0 -0.1 0.1 Gross horsepower0.0 -0.1 0.0 0.0 -0.1 0.0 0.0 0.0 0.1 Weight (1000 lbs)-4.6 -7.2 -2.0 -2.3 -5.0 0.4 -7.7 -12.5 -2.9 1/4 mile time0.5 -0.4 1.5 0.4 -0.7 1.6 1.6 -0.2 3.4 Example of data processing with multiple-response variables
Here we use truncated dataset with data from product test of two samples of chocolate sweets. 150 respondents tested two kinds of sweets (codenames: VSX123 and SDF546). Sample was divided into two groups (cells) of 75 respondents in each group. In cell 1 product VSX123 was presented first and then SDF546. In cell 2 sweets were presented in reversed order. Questions about respondent impressions about first product are in the block A (and about second tested product in the block B). At the end of the questionnaire there was a question about the preferences between sweets.
List of variables:
idRespondent IdcellFirst tested product (cell number)s2aAgea1_1-a1_6What did you like in these sweets? Multiple response. First tested producta22Overall quality. First tested productb1_1-b1_6What did you like in these sweets? Multiple response. Second tested productb22Overall quality. Second tested productc1Preferences
data(product_test) w = product_test # shorter name to save some keystrokes # here we recode variables from first/second tested product to separate variables for each product according to their cells # 'h' variables - VSX123 sample, 'p' variables - 'SDF456' sample # also we recode preferences from first/second product to true names # for first cell there are no changes, for second cell we should exchange 1 and 2. w = w %>% do_if(cell == 1, { recode(a1_1 %to% a1_6, other ~ copy) %into% (h1_1 %to% h1_6) recode(b1_1 %to% b1_6, other ~ copy) %into% (p1_1 %to% p1_6) recode(a22, other ~ copy) %into% h22 recode(b22, other ~ copy) %into% p22 c1r = c1 }) %>% do_if(cell == 2, { recode(a1_1 %to% a1_6, other ~ copy) %into% (p1_1 %to% p1_6) recode(b1_1 %to% b1_6, other ~ copy) %into% (h1_1 %to% h1_6) recode(a22, other ~ copy) %into% p22 recode(b22, other ~ copy) %into% h22 recode(c1, 1 ~ 2, 2 ~ 1, other ~ copy) %into% c1r }) %>% compute({ # recode age by groups age_cat = recode(s2a, lo %thru% 25 ~ 1, lo %thru% hi ~ 2) # count number of likes # codes 2 and 99 are ignored. h_likes = count_row_if(1 | 3 %thru% 98, h1_1 %to% h1_6) p_likes = count_row_if(1 | 3 %thru% 98, p1_1 %to% p1_6) }) # here we prepare labels for future usage codeframe_likes = num_lab(" 1 Liked everything 2 Disliked everything 3 Chocolate 4 Appearance 5 Taste 6 Stuffing 7 Nuts 8 Consistency 98 Other 99 Hard to answer ") overall_liking_scale = num_lab(" 1 Extremely poor 2 Very poor 3 Quite poor 4 Neither good, nor poor 5 Quite good 6 Very good 7 Excellent ") w = apply_labels(w, c1r = "Preferences", c1r = num_lab(" 1 VSX123 2 SDF456 3 Hard to say "), age_cat = "Age", age_cat = c("18 - 25" = 1, "26 - 35" = 2), h1_1 = "Likes. VSX123", p1_1 = "Likes. SDF456", h1_1 = codeframe_likes, p1_1 = codeframe_likes, h_likes = "Number of likes. VSX123", p_likes = "Number of likes. SDF456", h22 = "Overall quality. VSX123", p22 = "Overall quality. SDF456", h22 = overall_liking_scale, p22 = overall_liking_scale )Are there any significant differences between preferences? Yes, difference is significant.
# 'tab_mis_val(3)' remove 'hard to say' from vector w %>% tab_cols(total(), age_cat) %>% tab_cells(c1r) %>% tab_mis_val(3) %>% tab_stat_cases() %>% tab_last_sig_cases() %>% tab_pivot()#Total Age 18 - 25 26 - 35 Preferences VSX123 94.0 46.0 48.0 SDF456 50.0 22.0 28.0 Hard to say #Chi-squared p-value <0.05 (warn.) #Total cases 144.0 68.0 76.0 Further we calculate distribution of answers in the survey questions.
# lets specify repeated parts of table creation chains banner = w %>% tab_cols(total(), age_cat, c1r) # column percent with significance tab_cpct_sig = . %>% tab_stat_cpct() %>% tab_last_sig_cpct(sig_labels = paste0("<b>",LETTERS, "</b>")) # means with siginifcance tab_means_sig = . %>% tab_stat_mean_sd_n(labels = c("<b><u>Mean</u></b>", "sd", "N")) %>% tab_last_sig_means( sig_labels = paste0("<b>",LETTERS, "</b>"), keep = "means") # Preferences banner %>% tab_cells(c1r) %>% tab_cpct_sig() %>% tab_pivot()#Total Age Preferences 18 - 25 26 - 35 VSX123 SDF456 Hard to say A B A B C Preferences VSX123 62.7 65.7 60.0 100.0 SDF456 33.3 31.4 35.0 100.0 Hard to say 4.0 2.9 5.0 100.0 #Total cases 150 70 80 94 50 6 # Overall liking banner %>% tab_cells(h22) %>% tab_means_sig() %>% tab_cpct_sig() %>% tab_cells(p22) %>% tab_means_sig() %>% tab_cpct_sig() %>% tab_pivot()#Total Age Preferences 18 - 25 26 - 35 VSX123 SDF456 Hard to say A B A B C Overall quality. VSX123 Mean 5.5 5.4 5.6 5.3 5.8 A 5.5 Extremely poor Very poor Quite poor 2.0 2.9 1.2 3.2 Neither good, nor poor 10.7 11.4 10.0 14.9 B 2.0 16.7 Quite good 39.3 45.7 33.8 40.4 38.0 33.3 Very good 33.3 24.3 41.2 A 30.9 38.0 33.3 Excellent 14.7 15.7 13.8 10.6 22.0 16.7 #Total cases 150 70 80 94 50 6 Overall quality. SDF456 Mean 5.4 5.3 5.4 5.4 5.3 5.7 Extremely poor Very poor 0.7 1.2 1.1 Quite poor 2.7 4.3 1.2 2.1 4.0 Neither good, nor poor 16.7 20.0 13.8 18.1 14.0 16.7 Quite good 31.3 27.1 35.0 28.7 38.0 16.7 Very good 35.3 35.7 35.0 35.1 34.0 50.0 Excellent 13.3 12.9 13.8 14.9 10.0 16.7 #Total cases 150 70 80 94 50 6 # Likes banner %>% tab_cells(h_likes) %>% tab_means_sig() %>% tab_cells(mrset(h1_1 %to% h1_6)) %>% tab_cpct_sig() %>% tab_cells(p_likes) %>% tab_means_sig() %>% tab_cells(mrset(p1_1 %to% p1_6)) %>% tab_cpct_sig() %>% tab_pivot()#Total Age Preferences 18 - 25 26 - 35 VSX123 SDF456 Hard to say A B A B C Number of likes. VSX123 Mean 2.0 2.0 2.1 1.9 2.2 2.3 Likes. VSX123 Liked everything Disliked everything 3.3 1.4 5.0 4.3 2.0 Chocolate 34.0 38.6 30.0 35.1 32.0 33.3 Appearance 29.3 21.4 36.2 A 25.5 38.0 16.7 Taste 32.0 38.6 26.2 23.4 48.0 A 33.3 Stuffing 27.3 20.0 33.8 28.7 26.0 16.7 Nuts 66.7 72.9 61.3 69.1 60.0 83.3 Consistency 12.0 4.3 18.8 A 8.5 14.0 50.0 A B Other Hard to answer #Total cases 150 70 80 94 50 6 Number of likes. SDF456 Mean 2.0 2.0 2.1 2.0 2.0 2.0 Likes. SDF456 Liked everything Disliked everything 1.3 1.4 1.2 2.1 Chocolate 32.0 27.1 36.2 29.8 34.0 50.0 Appearance 32.0 35.7 28.7 34.0 30.0 16.7 Taste 39.3 42.9 36.2 36.2 44.0 50.0 Stuffing 27.3 24.3 30.0 31.9 20.0 16.7 Nuts 61.3 60.0 62.5 58.5 68.0 50.0 Consistency 10.0 5.7 13.8 11.7 6.0 16.7 Other 0.7 1.2 1.1 Hard to answer #Total cases 150 70 80 94 50 6 # below more complicated table where we compare likes side by side # Likes - side by side comparison w %>% tab_cols(total(label = "#Total| |"), c1r) %>% tab_cells(list(unvr(mrset(h1_1 %to% h1_6)))) %>% tab_stat_cpct(label = var_lab(h1_1)) %>% tab_cells(list(unvr(mrset(p1_1 %to% p1_6)))) %>% tab_stat_cpct(label = var_lab(p1_1)) %>% tab_pivot(stat_position = "inside_columns")#Total Preferences VSX123 SDF456 Hard to say Likes. VSX123 Likes. SDF456 Likes. VSX123 Likes. SDF456 Likes. VSX123 Likes. SDF456 Likes. VSX123 Likes. SDF456 Liked everything Disliked everything 3.3 1.3 4.3 2.1 2 Chocolate 34.0 32.0 35.1 29.8 32 34 33.3 50.0 Appearance 29.3 32.0 25.5 34.0 38 30 16.7 16.7 Taste 32.0 39.3 23.4 36.2 48 44 33.3 50.0 Stuffing 27.3 27.3 28.7 31.9 26 20 16.7 16.7 Nuts 66.7 61.3 69.1 58.5 60 68 83.3 50.0 Consistency 12.0 10.0 8.5 11.7 14 6 50.0 16.7 Other 0.7 1.1 Hard to answer #Total cases 150 150 94 94 50 50 6 6 We can save labelled dataset as *.csv file with accompanying R code for labelling.
write_labelled_csv(w, file filename = "product_test.csv")Or, we can save dataset as *.csv file with SPSS syntax to read data and apply labels.
write_labelled_spss(w, file filename = "product_test.csv")Labels support for base R
Variable label is human readable description of the variable. R supports rather long variable names and these names can contain even spaces and punctuation but short variables names make coding easier. Variable label can give a nice, long description of variable. With this description it is easier to remember what those variable names refer to. Value labels are similar to variable labels, but value labels are descriptions of the values a variable can take. Labeling values means we don’t have to remember if 1=Extremely poor and 7=Excellent or vice-versa. We can easily get dataset description and variables summary with
infofunction.The usual way to connect numeric data to labels in R is in factor variables. However, factors miss important features which the value labels provide. Factors only allow for integers to be mapped to a text label, these integers have to be a count starting at 1 and every value need to be labelled. Also, we can’t calculate means or other numeric statistics on factors.
With labels we can manipulate short variable names and codes when we analyze our data but in the resulting tables and graphs we will see human-readable text.
It is easy to store labels as variable attributes in R but most R functions cannot use them or even drop them.
expsspackage integrates value labels support into base R functions and into functions from other packages. Every function which internally converts variable to factor will utilize labels. Labels will be preserved during variables subsetting and concatenation. Additionally, there is a function (use_labels) which greatly simplify variable labels usage. See examples below.Getting and setting variable and value labels
First, apply value and variables labels to dataset:
library(expss) data(mtcars) mtcars = apply_labels(mtcars, mpg = "Miles/(US) gallon", cyl = "Number of cylinders", disp = "Displacement (cu.in.)", hp = "Gross horsepower", drat = "Rear axle ratio", wt = "Weight (1000 lbs)", qsec = "1/4 mile time", vs = "Engine", vs = c("V-engine" = 0, "Straight engine" = 1), am = "Transmission", am = c("Automatic" = 0, "Manual"=1), gear = "Number of forward gears", carb = "Number of carburetors" )In addition to
apply_labelswe have SPSS-stylevar_labandval_labfunctions:nps = c(-1, 0, 1, 1, 0, 1, 1, -1) var_lab(nps) = "Net promoter score" val_lab(nps) = num_lab(" -1 Detractors 0 Neutralists 1 Promoters ")We can read, add or remove existing labels:
var_lab(nps) # get variable label## [1] "Net promoter score"val_lab(nps) # get value labels## Detractors Neutralists Promoters ## -1 0 1# add new labels add_val_lab(nps) = num_lab(" 98 Other 99 Hard to say ") # remove label by value # %d% - diff, %n_d% - names diff val_lab(nps) = val_lab(nps) %d% 98 # or, remove value by name val_lab(nps) = val_lab(nps) %n_d% "Other"Additionaly, there are some utility functions. They can applied on one variable as well as on the entire dataset.
drop_val_labs(nps)## LABEL: Net promoter score ## VALUES: ## -1, 0, 1, 1, 0, 1, 1, -1drop_var_labs(nps)## VALUES: ## -1, 0, 1, 1, 0, 1, 1, -1 ## VALUE LABELS: ## -1 Detractors ## 0 Neutralists ## 1 Promoters ## 99 Hard to sayunlab(nps)## [1] -1 0 1 1 0 1 1 -1drop_unused_labels(nps)## LABEL: Net promoter score ## VALUES: ## -1, 0, 1, 1, 0, 1, 1, -1 ## VALUE LABELS: ## -1 Detractors ## 0 Neutralists ## 1 Promotersprepend_values(nps)## LABEL: Net promoter score ## VALUES: ## -1, 0, 1, 1, 0, 1, 1, -1 ## VALUE LABELS: ## -1 -1 Detractors ## 0 0 Neutralists ## 1 1 Promoters ## 99 99 Hard to sayThere is also
prepend_namesfunction but it can be applied only to data.frame.Labels with base R and ggplot2 functions
Base
tableand plotting with value labels:with(mtcars, table(am, vs))## vs ## am V-engine Straight engine ## Automatic 12 7 ## Manual 6 7with(mtcars, barplot( table(am, vs), beside = TRUE, legend = TRUE) )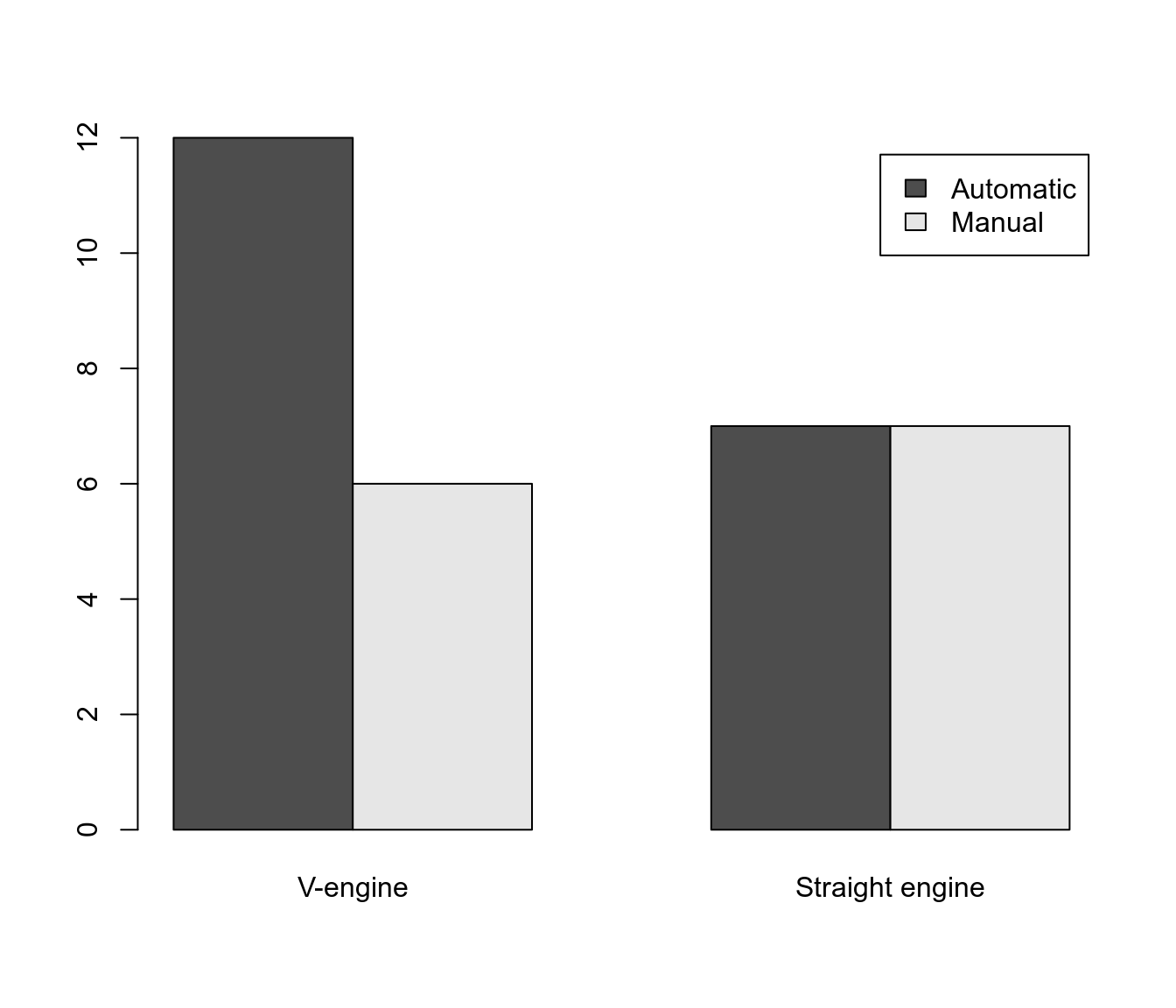
boxplot(mpg ~ am, data = mtcars)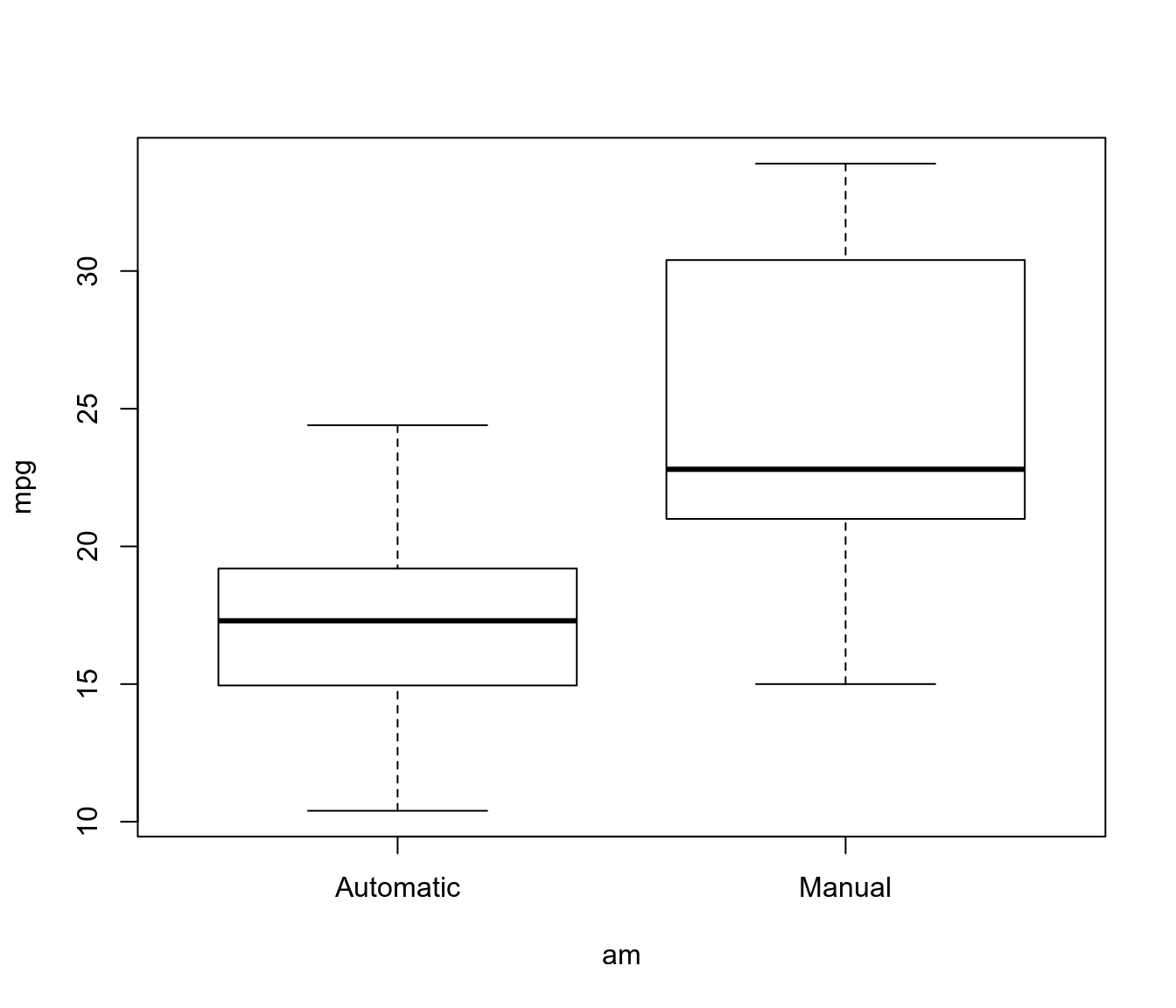
There is a special function for variables labels support -
use_labels. By now variables labels support available only for expression which will be evaluated inside data.frame.# table with dimension names use_labels(mtcars, table(am, vs))## Engine ## Transmission V-engine Straight engine ## Automatic 12 7 ## Manual 6 7# linear regression use_labels(mtcars, lm(mpg ~ wt + hp + qsec)) %>% summary## ## Call: ## lm(formula = `Miles/(US) gallon` ~ `Weight (1000 lbs)` + `Gross horsepower` + ## `1/4 mile time`) ## ## Residuals: ## LABEL: Miles/(US) gallon ## VALUES: ## -3.8591, -1.6418, -0.4636, 1.194, 5.6092 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 27.61053 8.41993 3.279 0.00278 ** ## `Weight (1000 lbs)` -4.35880 0.75270 -5.791 3.22e-06 *** ## `Gross horsepower` -0.01782 0.01498 -1.190 0.24418 ## `1/4 mile time` 0.51083 0.43922 1.163 0.25463 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 2.578 on 28 degrees of freedom ## Multiple R-squared: 0.8348, Adjusted R-squared: 0.8171 ## F-statistic: 47.15 on 3 and 28 DF, p-value: 4.506e-11And, finally,
ggplot2graphics with variables and value labels:library(ggplot2, warn.conflicts = FALSE) use_labels(mtcars, { # '..data' is shortcut for all 'mtcars' data.frame inside expression ggplot(..data) + geom_point(aes(y = mpg, x = wt, color = qsec)) + facet_grid(am ~ vs) })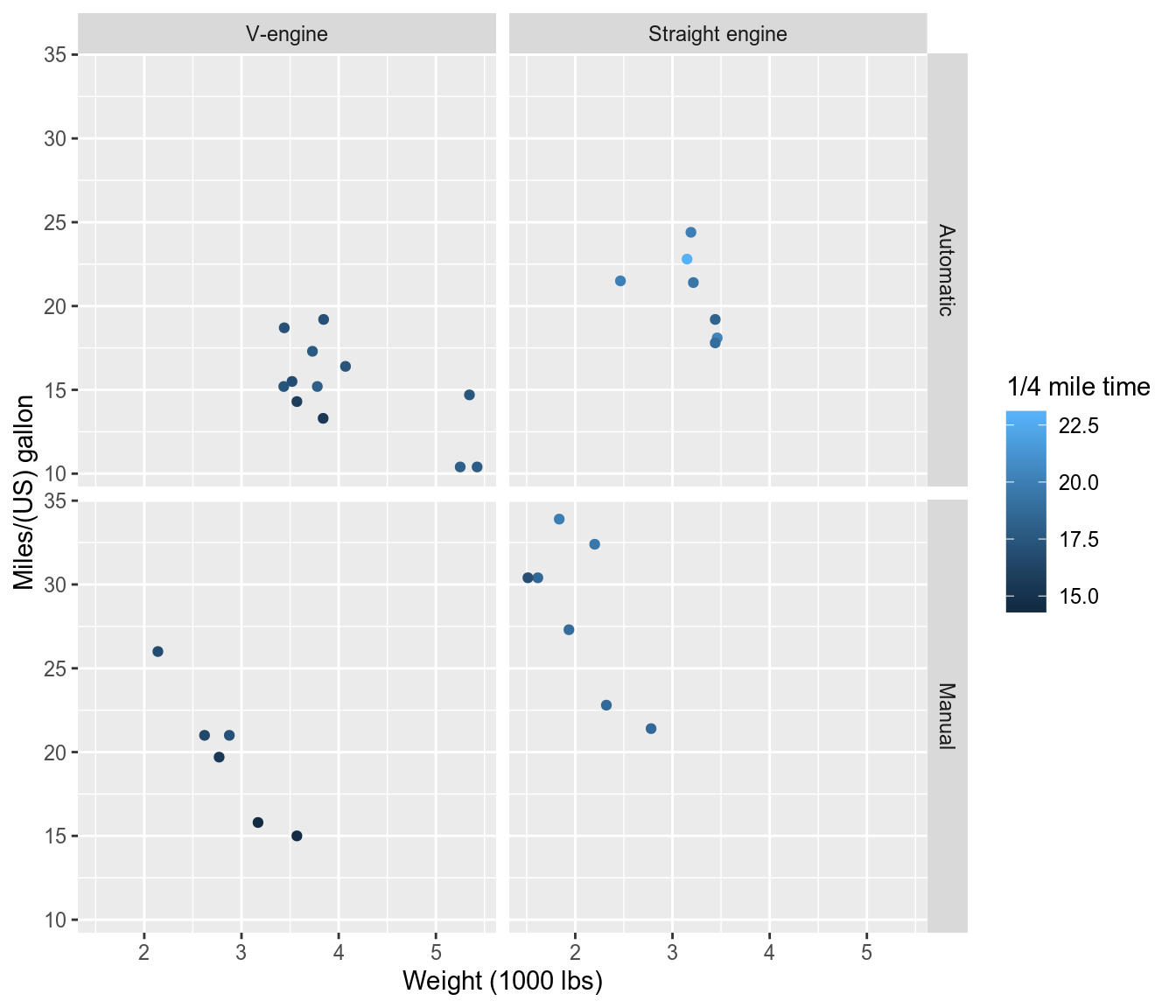
Extreme value labels support
We have an option for extreme values lables support:
expss_enable_value_labels_support_extreme(). With this optionfactor/as.factorwill take into account empty levels. However,uniquewill give weird result for labelled variables: labels without values will be added to unique values. That’s why it is recommended to turn off this option immediately after usage. See examples.We have label ‘Hard to say’ for which there are no values in
nps:nps = c(-1, 0, 1, 1, 0, 1, 1, -1) var_lab(nps) = "Net promoter score" val_lab(nps) = num_lab(" -1 Detractors 0 Neutralists 1 Promoters 99 Hard to say ")Here we disable labels support and get results without labels:
expss_disable_value_labels_support() table(nps) # there is no labels in the result## nps ## -1 0 1 ## 2 2 4unique(nps)## [1] -1 0 1Results with default value labels support - three labels are here but “Hard to say” is absent.
expss_enable_value_labels_support() # table with labels but there are no label "Hard to say" table(nps)## nps ## Detractors Neutralists Promoters ## 2 2 4unique(nps)## LABEL: Net promoter score ## VALUES: ## -1, 0, 1 ## VALUE LABELS: ## -1 Detractors ## 0 Neutralists ## 1 Promoters ## 99 Hard to sayAnd now extreme value labels support - we see “Hard to say” with zero counts. Note the weird
uniqueresult.expss_enable_value_labels_support_extreme() # now we see "Hard to say" with zero counts table(nps)## nps ## Detractors Neutralists Promoters Hard to say ## 2 2 4 0# weird 'unique'! There is a value 99 which is absent in 'nps' unique(nps)## LABEL: Net promoter score ## VALUES: ## -1, 0, 1, 99 ## VALUE LABELS: ## -1 Detractors ## 0 Neutralists ## 1 Promoters ## 99 Hard to sayReturn immediately to defaults to avoid issues:
expss_enable_value_labels_support()Labels are preserved during common operations on the data
There are special methods for subsetting and concatenating labelled variables. These methods preserve labels during common operations. We don’t need to restore labels on subsetted or sorted data.frame.
mtcarswith labels:str(mtcars)## 'data.frame': 32 obs. of 11 variables: ## $ mpg :Class 'labelled' num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ... ## .. .. LABEL: Miles/(US) gallon ## $ cyl :Class 'labelled' num 6 6 4 6 8 6 8 4 4 6 ... ## .. .. LABEL: Number of cylinders ## $ disp:Class 'labelled' num 160 160 108 258 360 ... ## .. .. LABEL: Displacement (cu.in.) ## $ hp :Class 'labelled' num 110 110 93 110 175 105 245 62 95 123 ... ## .. .. LABEL: Gross horsepower ## $ drat:Class 'labelled' num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ... ## .. .. LABEL: Rear axle ratio ## $ wt :Class 'labelled' num 2.62 2.88 2.32 3.21 3.44 ... ## .. .. LABEL: Weight (1000 lbs) ## $ qsec:Class 'labelled' num 16.5 17 18.6 19.4 17 ... ## .. .. LABEL: 1/4 mile time ## $ vs :Class 'labelled' num 0 0 1 1 0 1 0 1 1 1 ... ## .. .. LABEL: Engine ## .. .. VALUE LABELS [1:2]: 0=V-engine, 1=Straight engine ## $ am :Class 'labelled' num 1 1 1 0 0 0 0 0 0 0 ... ## .. .. LABEL: Transmission ## .. .. VALUE LABELS [1:2]: 0=Automatic, 1=Manual ## $ gear:Class 'labelled' num 4 4 4 3 3 3 3 4 4 4 ... ## .. .. LABEL: Number of forward gears ## $ carb:Class 'labelled' num 4 4 1 1 2 1 4 2 2 4 ... ## .. .. LABEL: Number of carburetorsMake subset of the data.frame:
mtcars_subset = mtcars[1:10, ]Labels are here, nothing is lost:
str(mtcars_subset)## 'data.frame': 10 obs. of 11 variables: ## $ mpg :Class 'labelled' num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ## .. .. LABEL: Miles/(US) gallon ## $ cyl :Class 'labelled' num 6 6 4 6 8 6 8 4 4 6 ## .. .. LABEL: Number of cylinders ## $ disp:Class 'labelled' num 160 160 108 258 360 ... ## .. .. LABEL: Displacement (cu.in.) ## $ hp :Class 'labelled' num 110 110 93 110 175 105 245 62 95 123 ## .. .. LABEL: Gross horsepower ## $ drat:Class 'labelled' num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ## .. .. LABEL: Rear axle ratio ## $ wt :Class 'labelled' num 2.62 2.88 2.32 3.21 3.44 ... ## .. .. LABEL: Weight (1000 lbs) ## $ qsec:Class 'labelled' num 16.5 17 18.6 19.4 17 ... ## .. .. LABEL: 1/4 mile time ## $ vs :Class 'labelled' num 0 0 1 1 0 1 0 1 1 1 ## .. .. LABEL: Engine ## .. .. VALUE LABELS [1:2]: 0=V-engine, 1=Straight engine ## $ am :Class 'labelled' num 1 1 1 0 0 0 0 0 0 0 ## .. .. LABEL: Transmission ## .. .. VALUE LABELS [1:2]: 0=Automatic, 1=Manual ## $ gear:Class 'labelled' num 4 4 4 3 3 3 3 4 4 4 ## .. .. LABEL: Number of forward gears ## $ carb:Class 'labelled' num 4 4 1 1 2 1 4 2 2 4 ## .. .. LABEL: Number of carburetorsInteraction with ‘haven’
To use
expsswithhavenyou need to loadexpssstrictly afterhaven(or other package with implemented ‘labelled’ class) to avoid conflicts. And it is better to useread_spsswith explict package specification:haven::read_spss. See example below.havenpackage doesn’t set ‘labelled’ class for variables which have variable label but don’t have value labels. It leads to labels losing during subsetting and other operations. We have a special function to fix this:add_labelled_class. Apply it to dataset loaded byhaven.# we need to load packages strictly in this order to avoid conflicts library(haven) library(expss) spss_data = haven::read_spss("spss_file.sav") # add missing 'labelled' class spss_data = add_labelled_class(spss_data)Export to Microsoft Excel
To export
expsstables to *.xlsx you need to install excellentopenxlsxpackage. To install it just type in the consoleinstall.packages("openxlsx"). On Windows system you may need also install RTools. It can be downloaded from CRAN: RTools.First we apply labels on the mtcars dataset and build simple table with caption.
library(expss) library(openxlsx) data(mtcars) mtcars = apply_labels(mtcars, mpg = "Miles/(US) gallon", cyl = "Number of cylinders", disp = "Displacement (cu.in.)", hp = "Gross horsepower", drat = "Rear axle ratio", wt = "Weight (lb/1000)", qsec = "1/4 mile time", vs = "Engine", vs = c("V-engine" = 0, "Straight engine" = 1), am = "Transmission", am = c("Automatic" = 0, "Manual"=1), gear = "Number of forward gears", carb = "Number of carburetors" ) mtcars_table = mtcars %>% calc_cro_cpct( cell_vars = list(cyl, gear), col_vars = list(total(), am, vs) ) %>% set_caption("Table 1") mtcars_tableTable 1 #Total Transmission Engine Automatic Manual V-engine Straight engine Number of cylinders 4 34.4 15.8 61.5 5.6 71.4 6 21.9 21.1 23.1 16.7 28.6 8 43.8 63.2 15.4 77.8 #Total cases 32 19 13 18 14 Number of forward gears 3 46.9 78.9 66.7 21.4 4 37.5 21.1 61.5 11.1 71.4 5 15.6 38.5 22.2 7.1 #Total cases 32 19 13 18 14 Then we create workbook and add worksheet to it.
wb = createWorkbook() sh = addWorksheet(wb, "Tables")Export - we should specify workbook and worksheet.
xl_write(mtcars_table, wb, sh)And, finally, we save workbook with table to the xlsx file.
saveWorkbook(wb, "table1.xlsx", overwrite = TRUE)Screenshot of the exported table:
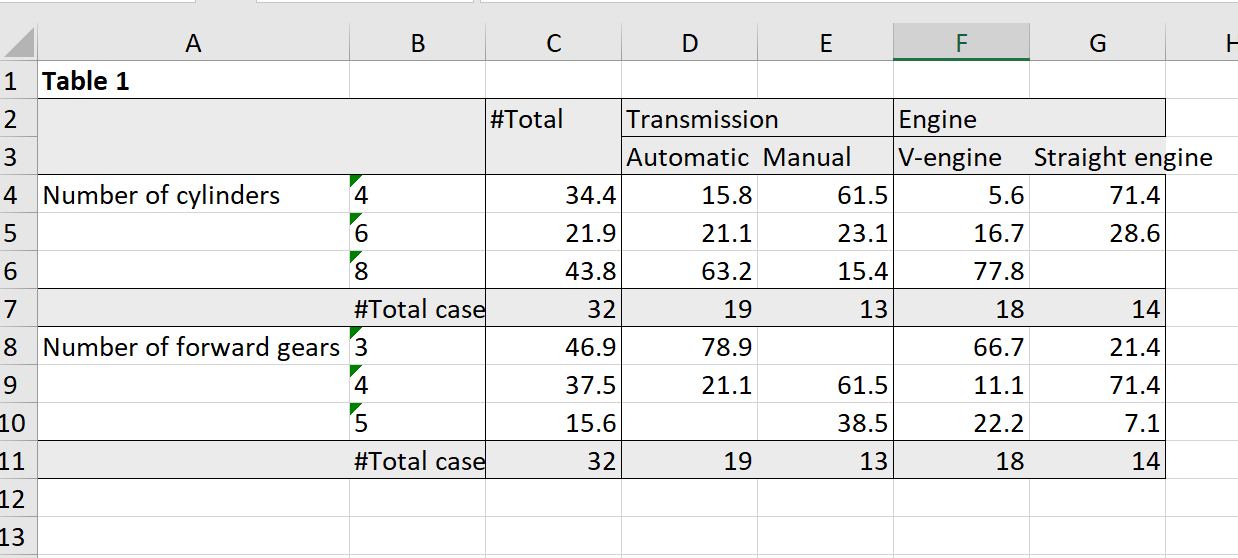
Automation of the report generation
First of all, we create banner which we will use for all our tables.
banner = calc(mtcars, list(total(), am, vs))Then we generate list with all tables. If variables have small number of discrete values we create column percent table. In other cases we calculate table with means. For both types of tables we mark significant differencies between groups.
list_of_tables = lapply(mtcars, function(variable) { if(length(unique(variable))<7){ cro_cpct(variable, banner) %>% significance_cpct() } else { # if number of unique values greater than seven we calculate mean cro_mean_sd_n(variable, banner) %>% significance_means() } })Create workbook:
wb = createWorkbook() sh = addWorksheet(wb, "Tables")Here we export our list with tables with additional formatting. We remove ‘#’ sign from totals and mark total column with bold. You can read about formatting options in the manual fro
xl_write(?xl_writein the console).xl_write(list_of_tables, wb, sh, # remove '#' sign from totals col_symbols_to_remove = "#", row_symbols_to_remove = "#", # format total column as bold other_col_labels_formats = list("#" = createStyle(textDecoration = "bold")), other_cols_formats = list("#" = createStyle(textDecoration = "bold")), )Save workbook:
saveWorkbook(wb, "report.xlsx", overwrite = TRUE)Screenshot of the generated report:
Excel functions translation guide
Let us consider Excel toy table:
A B C 1 2 15 50 2 1 70 80 3 3 30 40 4 2 30 40
Code for creating the same table in R:library(expss) w = text_to_columns(" a b c 2 15 50 1 70 80 3 30 40 2 30 40 ")wis the name of our table.IF
Excel:
IF(B1>60, 1, 0)R: Here we create new column with name
dwith results.ifelsefunction is from base R not from ‘expss’ package but included here for completeness.w$d = ifelse(w$b>60, 1, 0)If we need to use multiple transformations it is often convenient to use
computefunction. Insidecomputewe can put arbitrary number of the statements:w = compute(w, { d = ifelse(b>60, 1, 0) e = 42 abc_sum = sum_row(a, b, c) abc_mean = mean_row(a, b, c) })COUNTIF
Count 1’s in the entire dataset.
Excel:
COUNTIF(A1:C4, 1)R:
count_if(1, w)or
calculate(w, count_if(1, a, b, c))Count values greater than 1 in each row of the dataset.
Excel:
COUNTIF(A1:C1, ">1")R:
w$d = count_row_if(gt(1), w)or
w = compute(w, { d = count_row_if(gt(1), a, b, c) })Count values less than or equal to 1 in column A of the dataset.
Excel:
COUNTIF(A1:A4, "<=1")R:
count_col_if(le(1), w$a)Excel R <1 lt(1) <=1 le(1) <>1 ne(1) =1 eq(1) >=1 ge(1) >1 gt(1) SUM/AVERAGE
Sum all values in the dataset.
Excel:
SUM(A1:C4)R:
sum(w, na.rm = TRUE)Calculate average of each row of the dataset.
Excel:
AVERAGE(A1:C1)R:
w$d = mean_row(w)or
w = compute(w, { d = mean_row(a, b, c) })Sum values of column
Aof the dataset.Excel:
SUM(A1:A4)R:
sum_col(w$a)SUMIF/AVERAGEIF
Sum values greater than 40 in the entire dataset.
Excel:
SUMIF(A1:C4, ">40")R:
sum_if(gt(40), w)or
calculate(w, sum_if(gt(40), a, b, c))Sum values less than 40 in the each row of the dataset.
Excel:
SUMIF(A1:C1, "<40")R:
w$d = sum_row_if(lt(40), w)or
w = compute(w, { d = sum_row_if(lt(40), a, b, c) })Calculate average of
Bcolumn with columnAvalues less than 3.Excel:
AVERAGEIF(A1:A4, "<3", B1:B4)R:
mean_col_if(lt(3), w$a, data = w$b)or, if we want calculate means for both
bandccolumns:calculate(w, mean_col_if(lt(3), a, data = sheet(b, c)))VLOOKUP
Our dictionary for lookup:X Y 1 1 apples 2 2 oranges 3 3 peaches Code for creating the same dictionary in R:
dict = text_to_columns(" x y 1 apples 2 oranges 3 peaches ")Excel:
VLOOKUP(A1, $X$1:$Y$3, 2, FALSE)R:
w$d = vlookup(w$a, dict, 2)or, we can use column names:
w$d = vlookup(w$a, dict, "y")SPSS functions translation guide
COMPUTE
SPSS:
COMPUTE d = 1.R:
w$d = 1or, in the specific data.frame
w = compute(w, { d = 1 })There can be arbitrary number of statements inside
compute.IF
SPSS:
IF(a = 3) d = 2.R:
w = compute(w, { d = ifelse(a == 3, 2, NA) })or,
w = compute(w, { d = ifs(a == 3 ~ 2) })DO IF
SPSS:
DO IF (a>1). COMPUTE d = 4. END IF.R:
w = do_if(w, a>1, { d = 4 })There can be arbitrary number of statements inside
do_if.COUNT
SPSS:
COUNT cnt = a1 TO a5 (LO THRU HI).R:
cnt = count_row_if(lo %thru% hi, a1 %to% a5)SPSS:
COUNT cnt = a1 TO a5 (SYSMIS).R:
cnt = count_row_if(NA, a1 %to% a5)SPSS:
COUNT cnt = a1 TO a5 (1 THRU 5).R:
cnt = count_row_if(1 %thru% 5, a1 %to% a5)SPSS:
COUNT cnt = a1 TO a5 (1 THRU HI).R:
cnt = count_row_if(1 %thru% hi, a1 %to% a5)or,
cnt = count_row_if(ge(1), a1 %to% a5)SPSS:
COUNT cnt = a1 TO a5 (LO THRU 1).R:
cnt = count_row_if(lo %thru% 1, a1 %to% a5)or,
cnt = count_row_if (le(1), a1 %to% a5)SPSS:
COUNT cnt = a1 TO a5 (1 THRU 5, 99).R:
cnt = count_row_if(1 %thru% 5 | 99, a1 %to% a5)SPSS:
COUNT cnt = a1 TO a5(1,2,3,4,5, SYSMIS).R:
cnt = count_row_if(c(1:5, NA), a1 %to% a5)count_row_ifcan be used insidecompute.RECODE
SPSS:
RECODE V1 (0=1) (1=0) (2, 3=-1) (9=9) (ELSE=SYSMIS)R:
recode(v1) = c(0 ~ 1, 1 ~ 0, 2:3 ~ -1, 9 ~ 9, other ~ NA)SPSS:
RECODE QVAR(1 THRU 5=1)(6 THRU 10=2)(11 THRU HI=3)(ELSE=0).R:
recode(qvar) = c(1 %thru% 5 ~ 1, 6 %thru% 10 ~ 2, 11 %thru% hi ~ 3, other ~ 0)SPSS:
RECODE STRNGVAR ('A', 'B', 'C'='A')('D', 'E', 'F'='B')(ELSE=' ').R:
recode(strngvar) = c(c('A', 'B', 'C') ~ 'A', c('D', 'E', 'F') ~ 'B', other ~ ' ')SPSS:
RECODE AGE (MISSING=9) (18 THRU HI=1) (0 THRU 18=0) INTO VOTER.R:
voter = recode(age, NA ~ 9, 18 %thru% hi ~ 1, 0 %thru% 18 ~ 0) # or recode(age, NA ~ 9, 18 %thru% hi ~ 1, 0 %thru% 18 ~ 0) %into% voterrecodecan be used insidecompute.VARIABLE LABELS
SPSS:
VARIABLE LABELS a "Fruits" b "Cost" c "Price".R:
w = apply_labels(w, a = "Fruits", b = "Cost", c = "Price" )VALUE LABELS
SPSS:
VALUE LABELS a 1 "apples" 2 "oranges" 3 "peaches".R:
w = apply_labels(w, a = num_lab(" 1 apples 2 oranges 3 peaches ") )or,
val_lab(w$a) = num_lab(" 1 apples 2 oranges 3 peaches ")Tables
R:
fre(w$a) # Frequency of fruitsFruits Count Valid percent Percent Responses, % Cumulative responses, % apples 1 25 25 25 25 oranges 2 50 50 50 75 peaches 1 25 25 25 100 #Total 4 100 100 100 <NA> 0 0 cro_cpct(w$b, w$a) # Column percent of cost by fruitsFruits apples oranges peaches Cost 15 50 30 50 100 70 100 #Total cases 1 2 1 cro_mean(sheet(w$b, w$c), w$a) # Mean cost and price by fruitsFruits apples oranges peaches Cost 70 22.5 30 Price 80 45.0 40 'R' 카테고리의 다른 글
moonBook 패키지를 이용한 도넛 원그래프 작성하기 (0) 2018.12.27 plotly 패키지를 이용한 원그래프 그리기 (0) 2018.12.27 특정한 데이터 유형을 갖는 열을 추출하기 (0) 2018.12.26 type() 함수와 mode() 함수의 차이에 대한 이해 (0) 2018.12.16 Jupyter Notebook에 R연동하기 (0) 2018.12.11
